“Over the past decade, we have focused on uncovering the ecological and evolutionary consequences of the extensive genomic heterogeneity observed in natural prokaryotic populations.”
Microbial Genomics and Evolution

“Over the past decade, we have focused on uncovering the ecological and evolutionary consequences of the extensive genomic heterogeneity observed in natural prokaryotic populations. By leveraging the vast information generated through comparative genomics, transcriptomics, and metagenomics, we have made significant strides in characterizing the genomic diversity of key microbial groups inhabiting the ocean’s epipelagic zone. Recent advancements, such as long-read metagenomics and single-cell sequencing, allowed us to investigate this diversity at unprecedented resolution. For instance, we have published several studies on marine streamlined bacterial groups such as SAR11, HIMB59, SAR116, SAR86, and the two marine pelagic Actinobacteriota lineages: Candidatus Actinomarinales and the Acidimicrobiales. These studies, grounded in an ecogenomic perspective, have revealed critical insights into their genomic variability and ecological roles. Additionally, we have expanded our understanding of viral diversity associated with these microbes, which has improved our understanding of the interactions between microbial hosts and their associated viruses in marine ecosystems.
Our work highlights the evolutionary strategies of microbes with streamlined genomes to adapt and thrive under the oligotrophic conditions that dominate surface ocean waters. At the individual level, these microbes experience evolutionary pressures that optimize nutrient use through gene loss, as the «streamlining theory” proposes. At the population level, however, these species serve as reservoirs of extensive genetic diversity. Subpopulations coexist and share public goods, preserving this genetic richness within natural populations. This dynamic not only maximizes resource utilization but also fosters a form of evolutionary cooperation.”
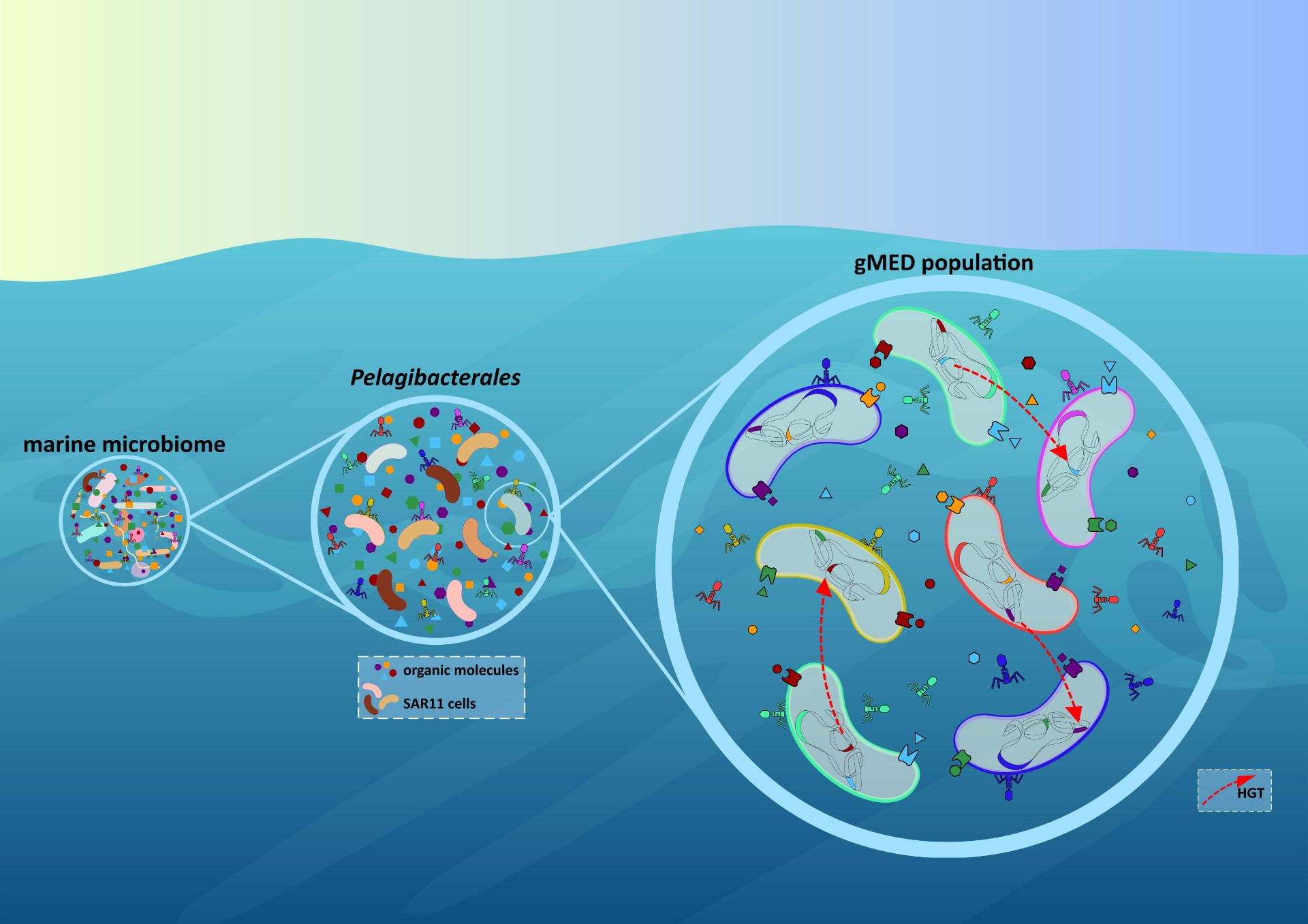
Population-level strategies that drive the ecological and evolutionary success of SAR11 in nutrient-limited marine environments. In oligotrophic ocean surface waters, where organic and inorganic nutrients are scarce, Pelagibacterales (SAR11 clade) has evolved under the principles of streamlining theory, including a reduced surface-to-volume ratio, resulting in very small cell sizes, as well as compact, highly efficient genomes. Beyond these individual adaptations, our findings reveal a complex population-level strategy that enhances evolutionary success. Conserved genomic location of variable regions facilitate homologous recombination of small DNA fragments, preserving core functions while maintaining functional redundancy within genomic islands (GIs). This redundancy sustains a vast environmental gene pool, particularly for nutrient transport, enabling broad substrate assimilation. Additionally, accessory genes within GIs enhance metabolic versatility, expanding the range of utilizable nutrients. As a result, SAR11 populations functions as a dynamic genetic reservoir, fostering polyclonality and the exchange of public goods (flexome), ensuring adaptability and stability within natural populations. (https://doi.org/10.1186/s40168-025-02037-6).
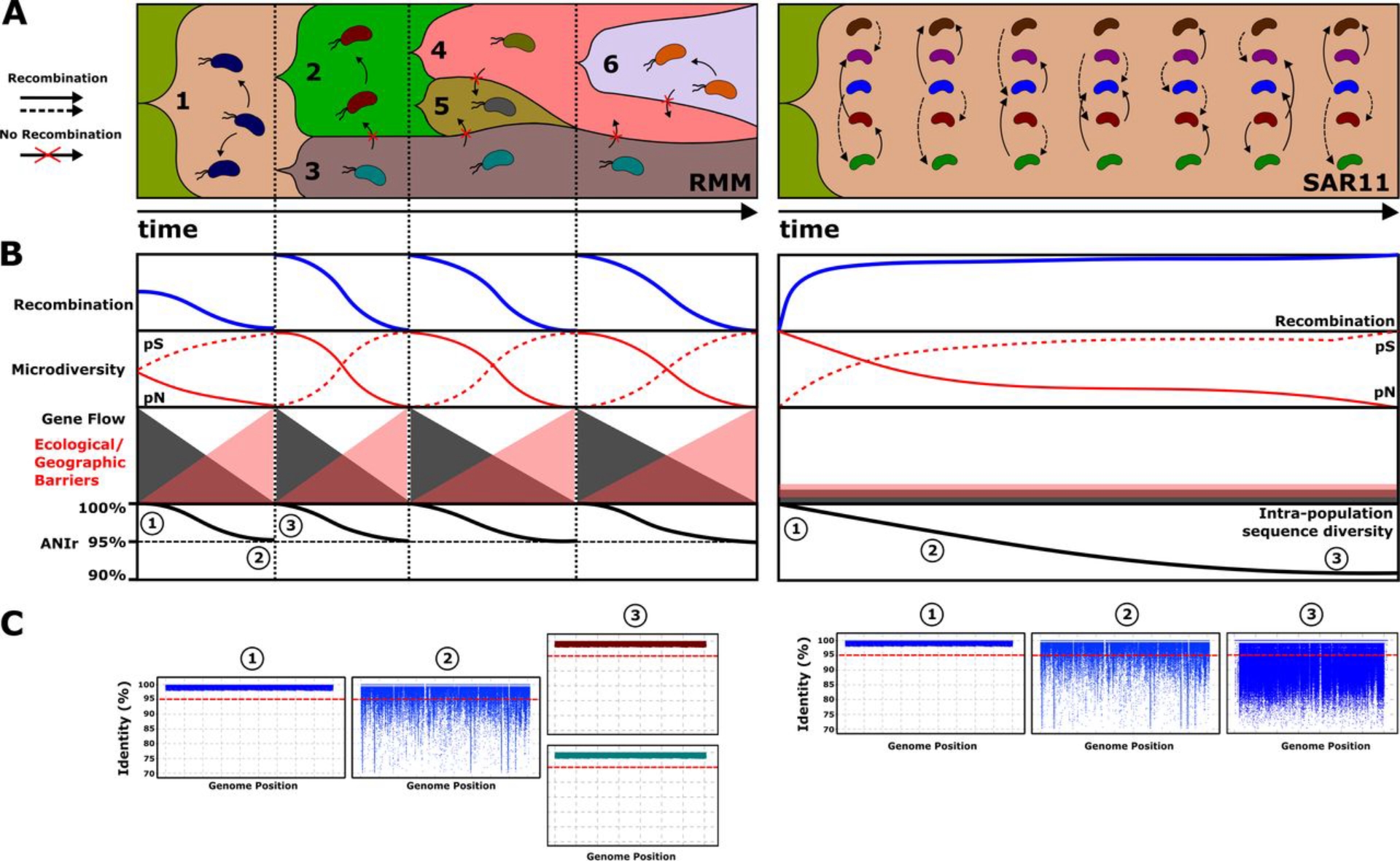
Evolutionary model for reference marine microbes (RMM) and SAR11 populations. RMM evolution is characterized by “hard” selective sweeps that reduce genomic diversity, and only a single adaptive lineage arises from the population. However, SAR11 evolution is characterized by “soft” sweeps, where multiple beneficial mutations contribute to an adaptive substitution, which are dispersed by the population, contributing to an increase in genetic diversity. (A) Relative abundances of the populations (y axis) and evolution over time (x axis). (B) Dynamics of different evolutionary parameters over time, where each vertical dotted line differentiates different diversification events in RMM, while in SAR11 populations, the same structure is maintained over time. (C) Recruitment plot showing intrapopulation sequence diversity. The red dashed line indicates the species threshold (95%). Different colors indicate different populations. (doi: 10.1128/mSystems.00605-20).
Microbial Genomics and Evolution
VISITING ADDRESS
Universidad Miguel Hernández. Edificio Muhammad Al-Shafra. Dpto. Producción Vegetal y Microbiología.
Ctra. Alicante-Valencia N-332, s/n. San Juan de Alicante, Alicante (Spain) – CP 03550
